// additive_log_ratio.stan
functions {
matrix compute_alr_probs(int n, int K, int p, matrix X, matrix beta) {
matrix[n, K] probs;
matrix[n, K - 1] expXbeta = exp(X * beta);
for (i in 1:n) {
real sum_i = sum(expXbeta[i, ]);
for (j in 1:K) {
if (j < K) {
probs[i, j] = expXbeta[i, j] / (1 + sum_i);
}
if (j == K) probs[i, j] = 1 - sum(probs[i, 1:(K - 1)]);
}
}
return probs
}
}
data {
int<lower = 1> K;
int<lower = 1> n;
int<lower = 1> p;
array[n] int<lower = 1, upper = K> Y;
matrix[n, p] X;
matrix[n, K] delta;
}
parameters {
matrix[p, K - 1] beta;
}
model {
matrix[n, K] probs = compute_alr_probs(n, K, p, X, beta);
for (i in 1:n) {
for (j in 1:K) {
target += delta[i, j] * log(probs[i, j]);
}
}
target += normal_lpdf(to_vector(beta) | 0, 10);
}Multiclass Classification
Feb 20, 2025
Review of last lecture
On Tuesday, we learned about classification using logistic regression.
Today, we will focus on multiclass classification: multinomial regression, ordinal regression.
Multiclass regression
Often times one encounters an outcome variable that is nominal and has more than two categories.
If there is no inherent rank or order to the variable, we can use multinomial regression. Examples include:
gender (male, female, non-binary),
blood type (A, B, AB, O).
If there is an order to the variable, we can use ordinal regression. Examples include:
stages of cancer (stage I, II, III, IV),
pain level (mild, moderate, severe).
Multinomial random variable
Assume an outcome
The likelihood in multinomial regression can be written as the following categorical likelihood,
Since
Log-linear regression
- One way to motivate multinomial regression is through a log-linear specification:
Finding the normalizing constant
- We know that,
Multinomial probabilities
- Thus, we have the following,
This function is called the softmax function.
Unfortunately, this specification is not identifiable.
Identifiability issue
- We can add a vector
- A common solution is to set:
Updating the probabilities
Using the identifiability constraint of
How to interpret the
Deriving the additive log ratio model
- Using our specification of the probabilities, it can be seen that,
Additive log ratio model
- If outcome
This formulation is called additive log ratio.
Getting back to the likelihood
- The log-likelihood can be written as,
The
As Bayesians, we only need to specify priors for
Multinomial regression in Stan
- Hard coding the likelihood.
Multinomial regression in Stan
- Non-identifiable version.
// multi_logit_bad.stan
data {
int<lower = 1> K;
int<lower = 1> n;
int<lower = 1> p;
array[n] int<lower = 1, upper = K> Y;
matrix[n, p] X;
}
parameters {
matrix[p, K] beta;
}
model {
matrix[n, K] Xbeta = X * beta;
for (i in 1:n) {
target += categorical_logit_lpmf(Y[i] | Xbeta[i]')
}
target += normal_lpdf(to_vector(beta) | 0, 10);
}Multinomial regression in Stan
- Zero identifiability constraint.
// multi_logit.stan
data {
int<lower = 1> K;
int<lower = 1> n;
int<lower = 1> p;
array[n] int<lower = 1, upper = K> Y;
matrix[n, p] X;
}
transformed data {
vector[p] zeros = rep_vector(0, p);
}
parameters {
matrix[p, K - 1] beta_raw;
}
transformed parameters {
matrix[p, K] beta = append_col(beta_raw, zeros);
}
model {
matrix[n, K] Xbeta = X * beta;
for (i in 1:n) {
target += categorical_logit_lpmf(Y[i] | Xbeta[i]')
}
target += normal_lpdf(to_vector(beta) | 0, 10);
}
generated quantities {
matrix[p, K - 1] ors = exp(beta_raw);
vector[n] Y_pred;
vector[n] log_lik;
for (i in 1:n) {
Y_pred[i] = categorical_logit_rng(Xbeta[i]');
log_lik[i] = categorical_logit_lpmf(Y[i] | Xbeta[i]');
}
}Ordinal regression
Let
- The likelihood in ordinal regression is identical to the one from multinomial regression,
- We need to add additional constraints that guarantee ordinality.
Proportional odds assumption
The odds of being less than or equal to a particular category can be defined as,
Not defined for
Proportional odds regression
The log odds can then be modeled as follows,
Why
Understanding the probabilities
- One can solve for
The individual probabilities are then given by,
A latent variable representation
- Define a latent variable,
CDF:
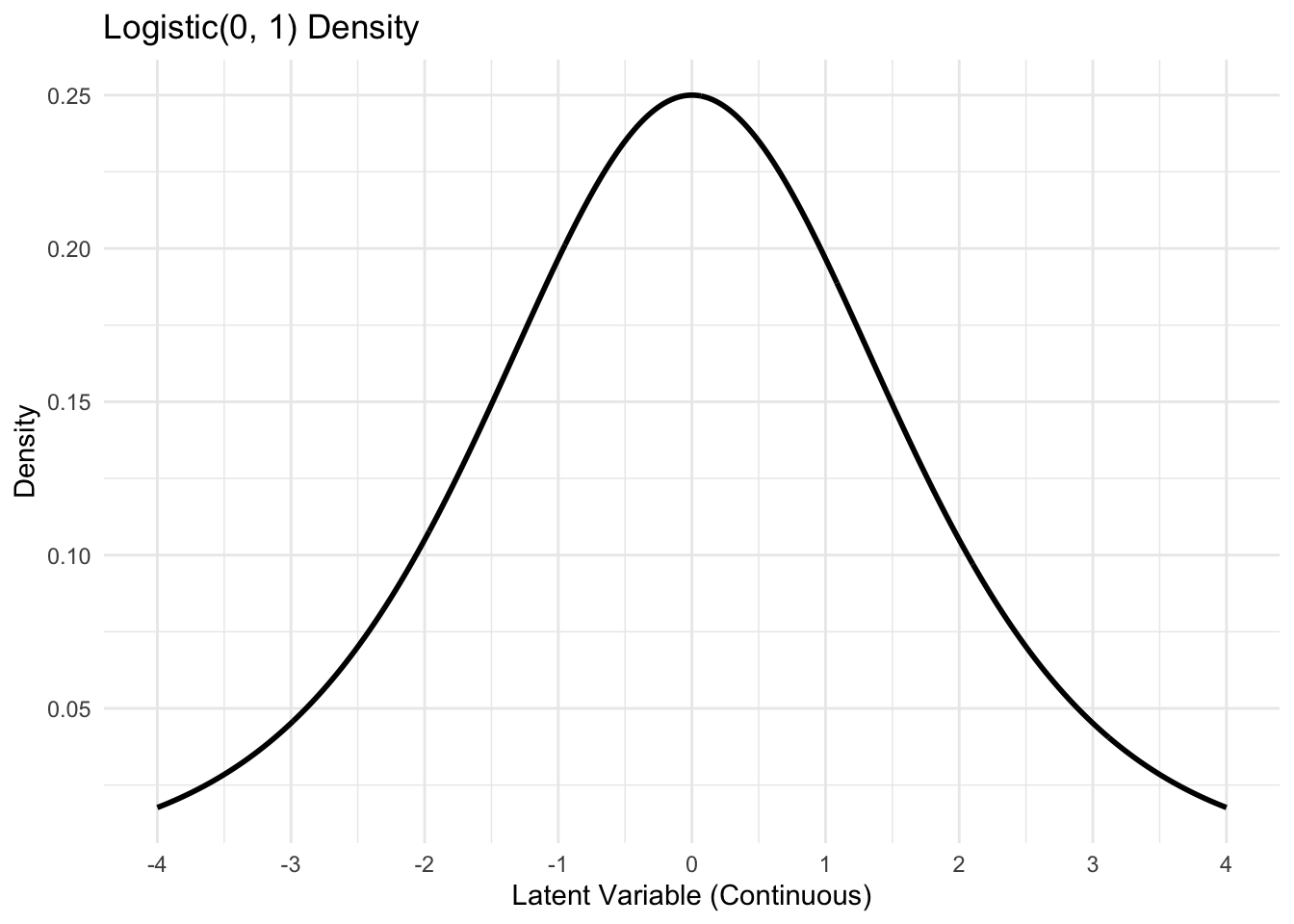
Visualizing the latent process
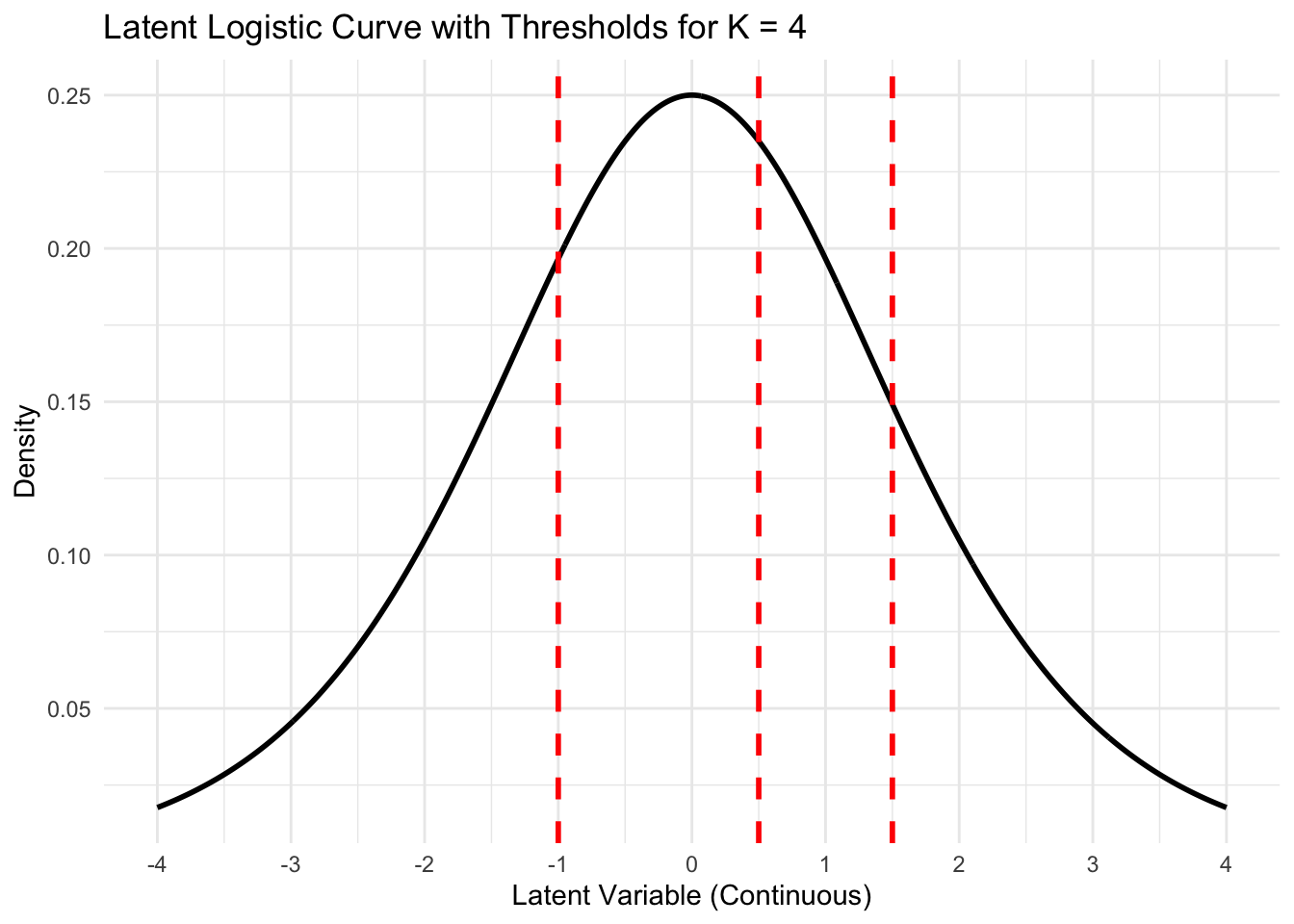
Adding thresholds (
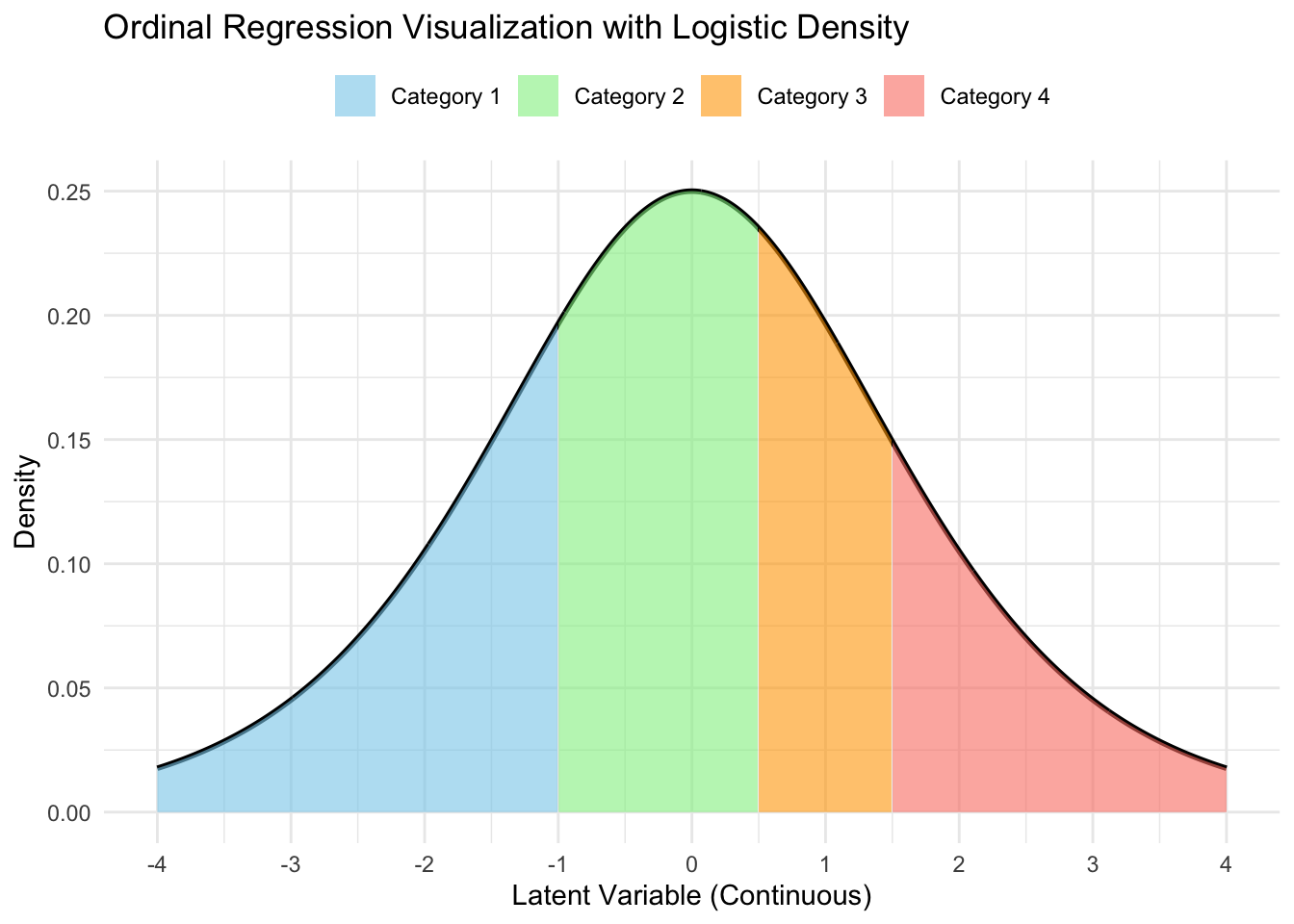
Getting category probabilities
A latent variable representation
Define a set of
Our ordinal random variable can be generated as,
Equivalence of the two specifications
Probabilities under the latent specification:
Equivalency:
Ordinal regression using Stan
// ordinal.stan
data {
int<lower = 2> K;
int<lower = 0> n;
int<lower = 1> p;
int<lower = 1, upper = K> Y[n];
matrix[n, p] X;
}
parameters {
vector[p] beta;
ordered[K - 1] alpha;
}
model {
target += ordered_logistic_glm_lpmf(Y | X, beta, alpha);
target += normal_lpdf(beta | 0, 10);
target += normal_lpdf(alpha | 0, 10);
}
generated quantities {
vector[p] ors = exp(beta);
vector[n] Y_pred;
vector[n] log_lik;
for (i in 1:n) {
Y_pred[i] = ordered_logistic_rng(X[i, ] * beta, alpha);
log_lik[i] = ordered_logistic_glm_lpmf(Y[i] | X[i, ], beta, alpha);
}
}Enforcing order in the cutoffs
In Stan, when you define a parameter as
ordered[K-1] alpha;, the values ofalphaare automatically constrained to be strictly increasing.This transformation ensures that the
alphavalues follow the required order, i.e.,alpha[1] < alpha[2] < ... < alpha[K-1].Stan doesn’t sample
alphadirectly but instead works with an unconstrained parameter vector, which we will callgamma.
Enforcing order in the cutoffs
Here,
gammarepresents a vector of independent, unconstrained variables, and the transformation ensures thatalphais strictly increasing by construction.Luckily we can use ordered, since Stan takes care of this (including the Jacobian) in the background.
Prepare for next class
Work on HW 03.
Complete reading to prepare for next Tuesday’s lecture
Tuesday’s lecture: Missing data