u = center(observed and prediction locations)
S = box size (u)
max_iter = 30
# initialization
j = 0
check = FALSE
rho = 0.5*S # the practical paper recommends setting the initial guess of rho to be 0.5 to 1 times S
c = c(rho/S) # minimum c given rho and S
m = m(c,rho/S) # minimum m given c, and rho/S
L = c*S
diagnosis = logical(max_iter) # store checking results for each iteration
while (!check & j<=max_iter){
fit = runHSGP(L,m) # stan run
j = j + 1
rho_hat = mean(fit$rho) # obtain fitted value for rho
# check the fitted is larger than the minimum rho that can be well approximated
diagnosis[j] = (rho_hat + 0.01 >= rho)
if (j==1) {
if (diagnosis[j]){
# if the diagnosis check is passed, do one more run just to make sure
m = m + 2
c = c(rho_hat/S)
rho = rho(m,c,S)
} else {
# if the check failed, update our knowledge about rho
rho = rho_hat
c = c(rho/S)
m = m(c,rho/S)
}
} else {
if (diagnosis[j] & diagnosis[j-2]){
# if the check passed for the last two runs, we finish tuning
check = TRUE
} else if (diagnosis[j] & !diagnosis[j-2]){
# if the check failed last time but passed this time, do one more run
m = m + 2
c = c(rho_hat/S)
rho = rho(m,c,S)
} else if (!diagnosis[j]){
# if the check failed, update our knowledge about rho
rho = rho_hat
c = c(rho/S)
m = m(c,rho/S)
}
}
L = c*S
}Scalable Gaussian Processes #2
Apr 03, 2025
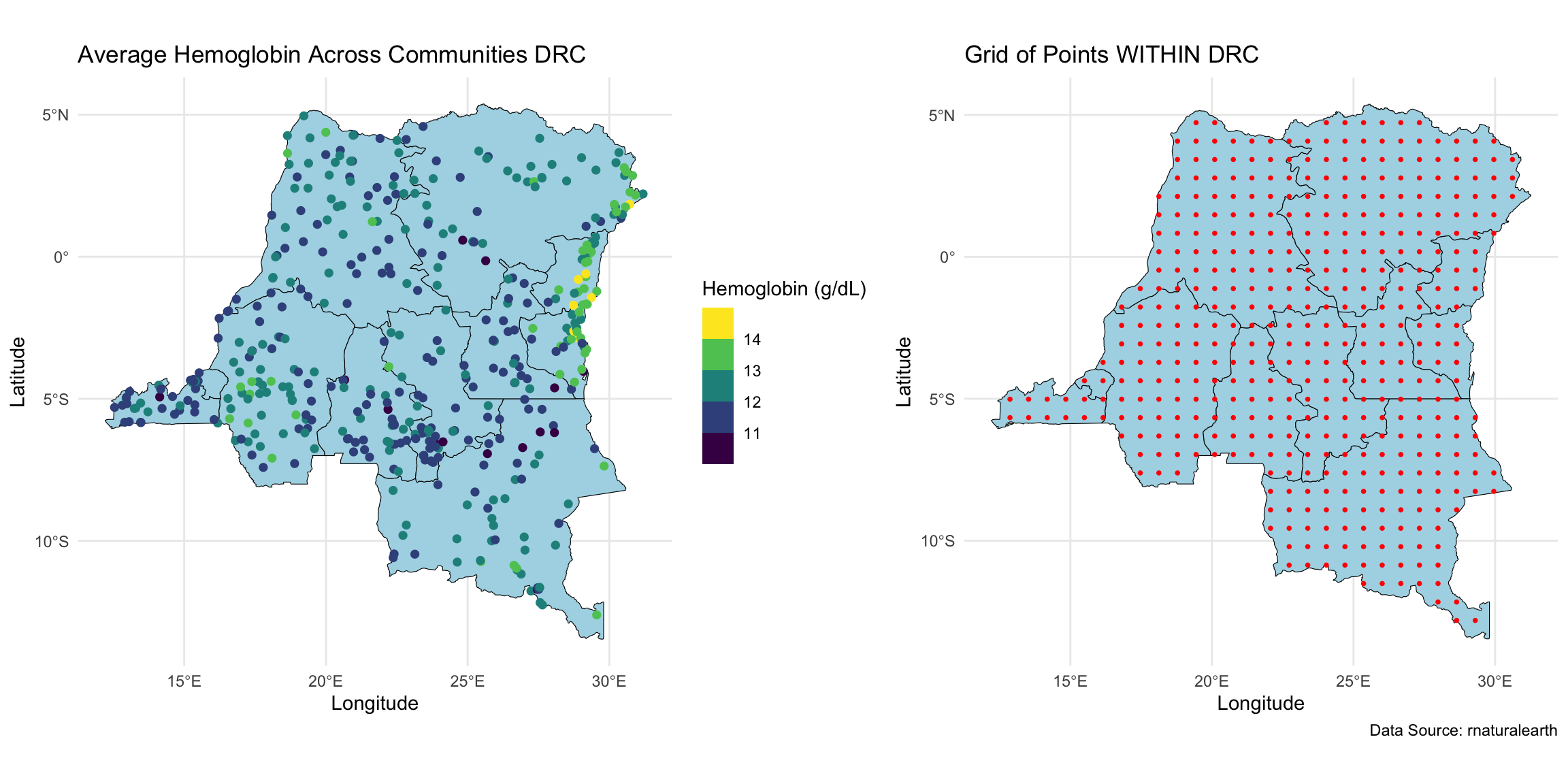
Geospatial analysis on hemoglobin dataset
We wanted to perform geospatial analysis on a dataset with ~8,600 observations at ~500 locations, and make predictions at ~440 locations on a grid.
Geospatial model
We specify the following model:
Review of the last lecture
Gaussian process (GP) is not scalable as it requires
Introduced HSGP, a Hilbert space low-rank approximation method for GP.
Model reparameterization under HSGP.
Bayesian model fitting and kriging under HSGP.
HSGP parameters
Solin and Särkkä (2020) showed that HSGP approximation can be made arbitrarily accurate as
Our goal:
- Minimize the run time while maintaining reasonable approximation accuracy.
- Find minimum
Note: we treat estimation of the GP magnitude parameter
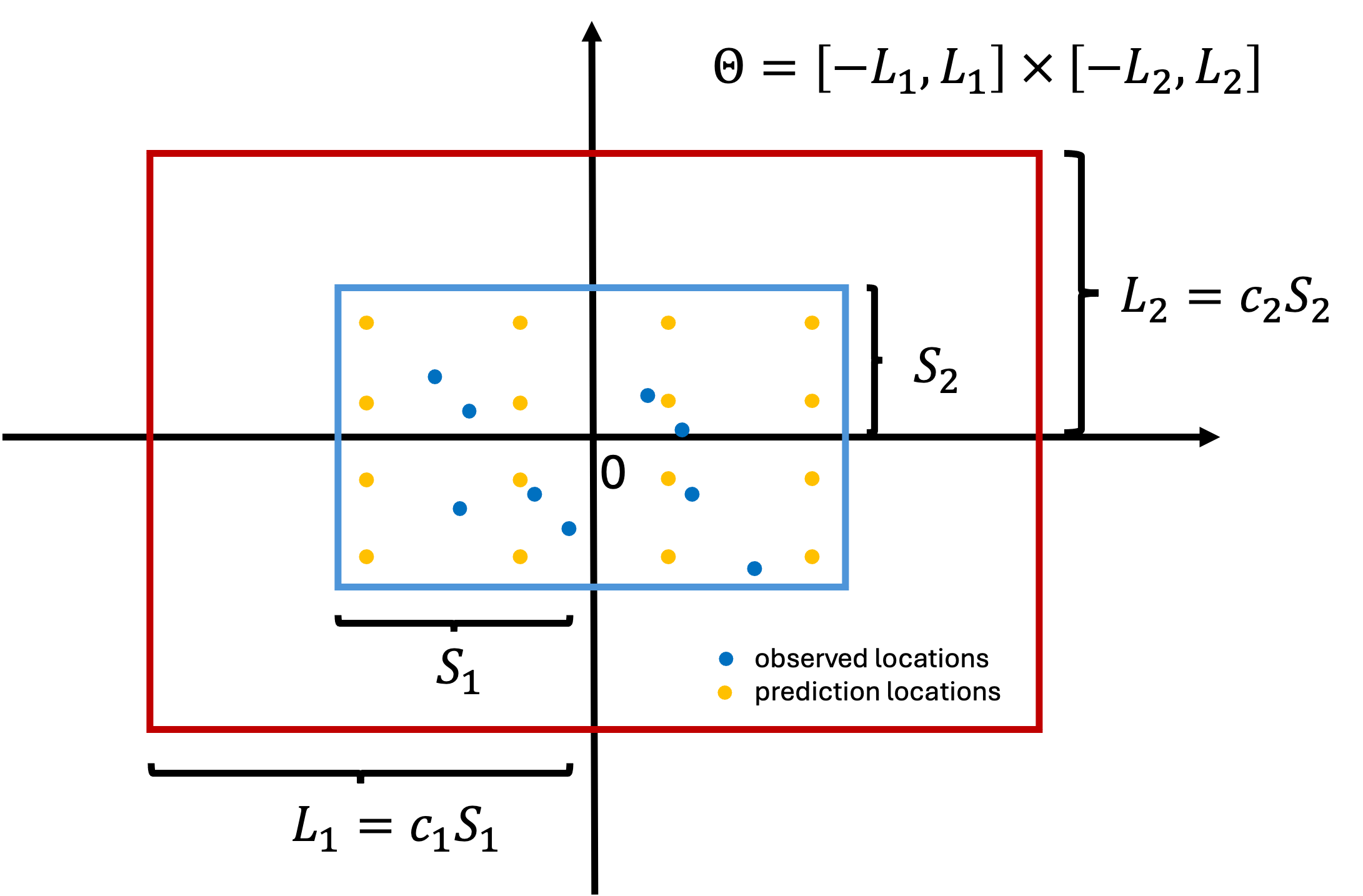
HSGP approximation box
Due to the design of HSGP, the approximation is less accurate near the boundaries of
- Suppose all the coordinates are centered. Let
- We want the box to be large enough to ensure good boundary accuracy. Let
HSGP approximation box and
How much the approximation accuracy deteriorates towards the boundaries depends on smoothness of the true surface.
- the larger the length scale
HSGP approximation box and

The larger the box,
- the more basis functions we need for the same level of overall accuracy,
- hence higher run time.
Zooming out doesn’t simplify the problem
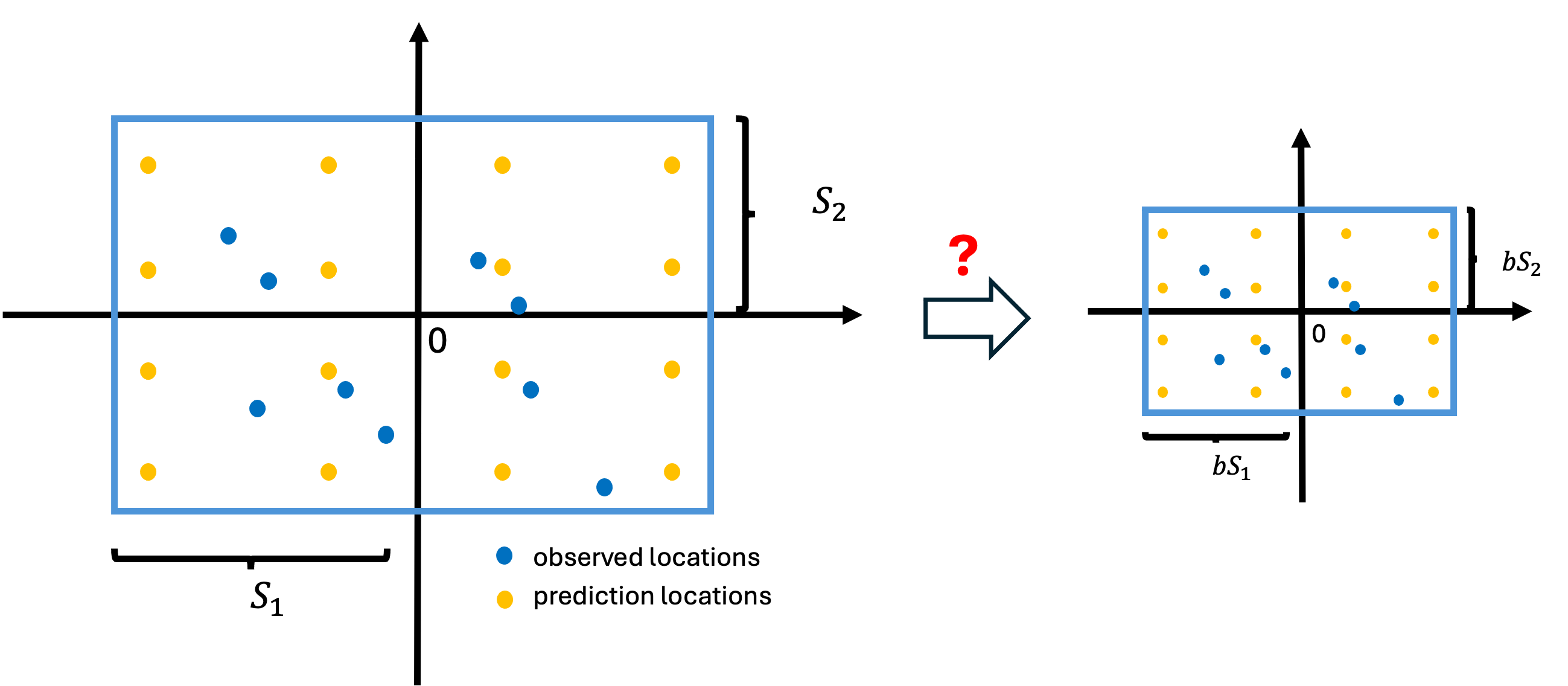
- If we scale the coordinates by a constant
- We can effectively think of the length scale parameter as
HSGP basis functions
The total number of basis functions
- The higher the
Relationship between
Let’s quickly recap. For simplicity, let
- As
- As
- As
Empirical functional form
Still assuming
- given
- Riutort-Mayol et al. (2023) used extensive simulations to obtain an empirical function form of
Empirical functional form
- Notice the linear proportionality between
- For a given
- From
Question
BUT, in real applications, we do not know
So how to make use of
An iterative algorithm
Pseudo-codes for HSGP parameter tuning assuming
HSGP implementation codes
Please clone the repo for AE 09 for HSGP implementation codes.
Side notes on HSGP implementation
A few random things to keep in mind for implementation in practice:
- If your HSGP run is suspiciously VERY slow, check the number of basis functions being used in the run and make sure it is reasonable.
- Check whether
- Because HSGP is a low-rank approximation method, the GP magnitude parameter
stancodes for parametertau_adj. - If
Side notes on HSGP implementation
A few random things to keep in mind for implementation in practice:
It is possible to use different length scale parameters for each dimension. See demo codes here for examples.
The iterative algorithm described in Riutort-Mayol et al. (2023) (i.e., pseudo-codes on slide 15) can be further improved:
- it sometimes stops at a place.
- it sometimes runs into a circular loop. See AE 09
stancodes for one possible fix. - it errs on the safe side and only changes
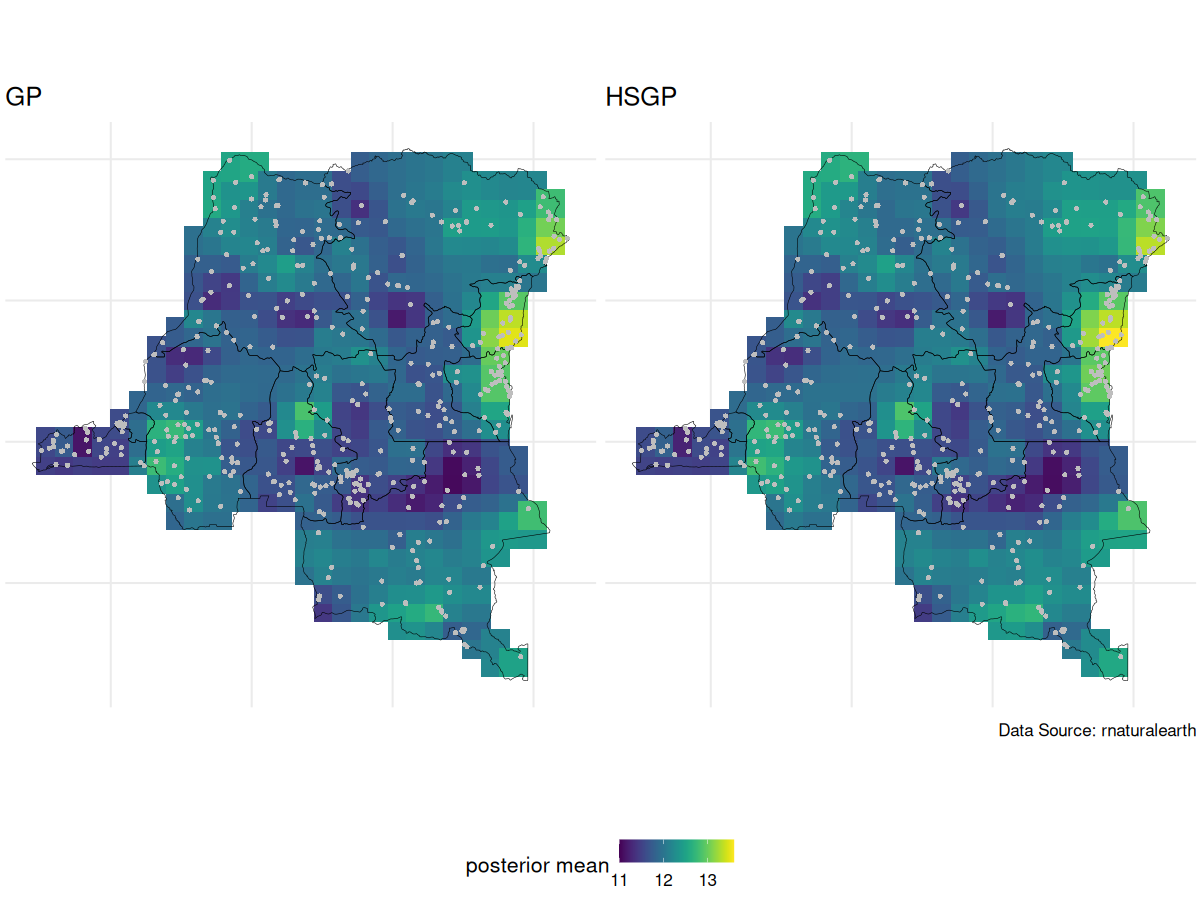
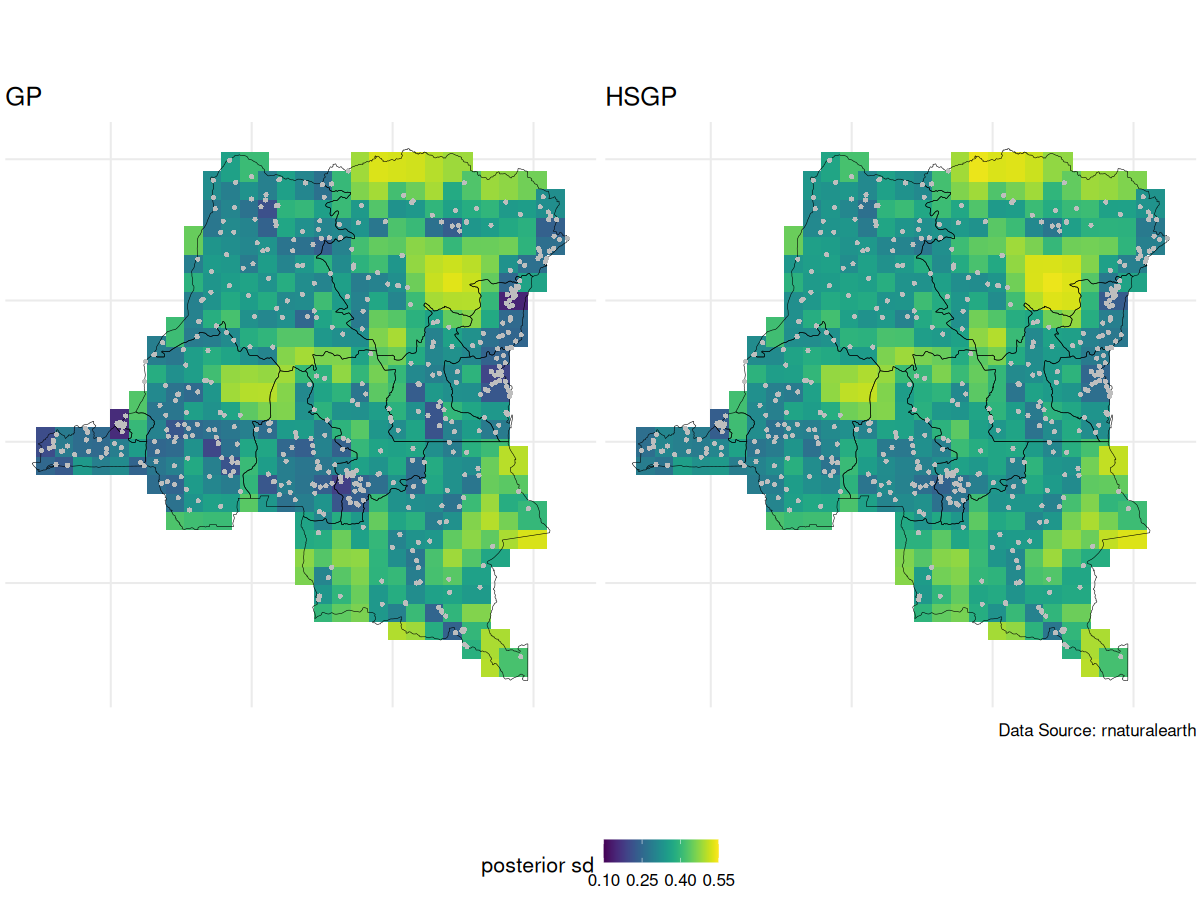
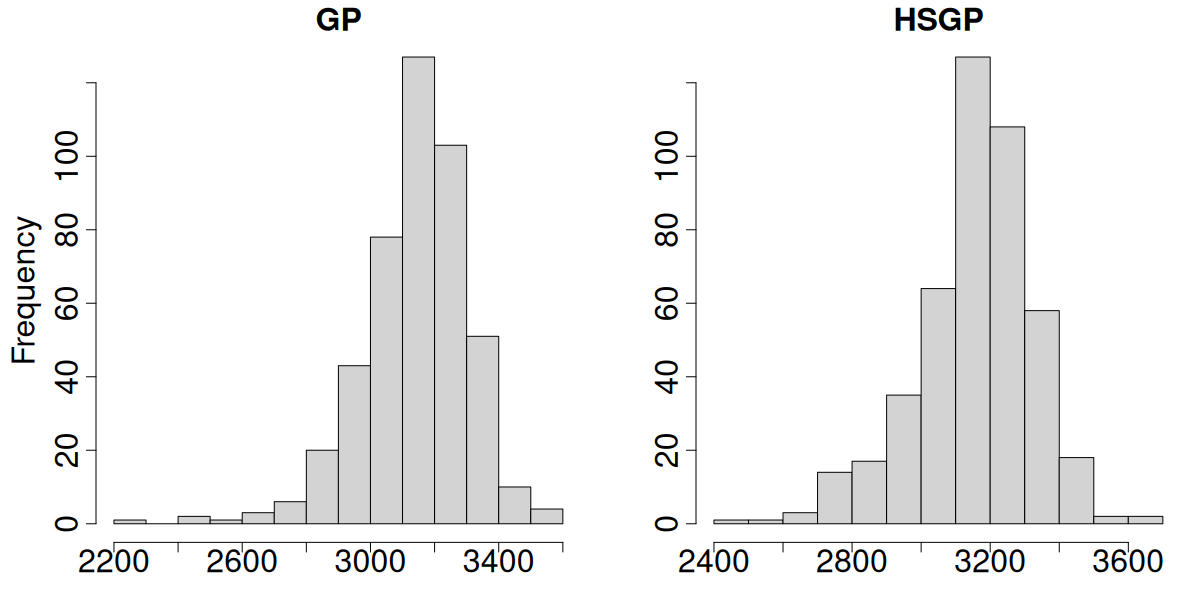
Due to identifiability issues, we always look at the spatial intercept
GP vs HSGP spatial intercept posterior mean
GP vs HSGP spatial intercept posterior SD
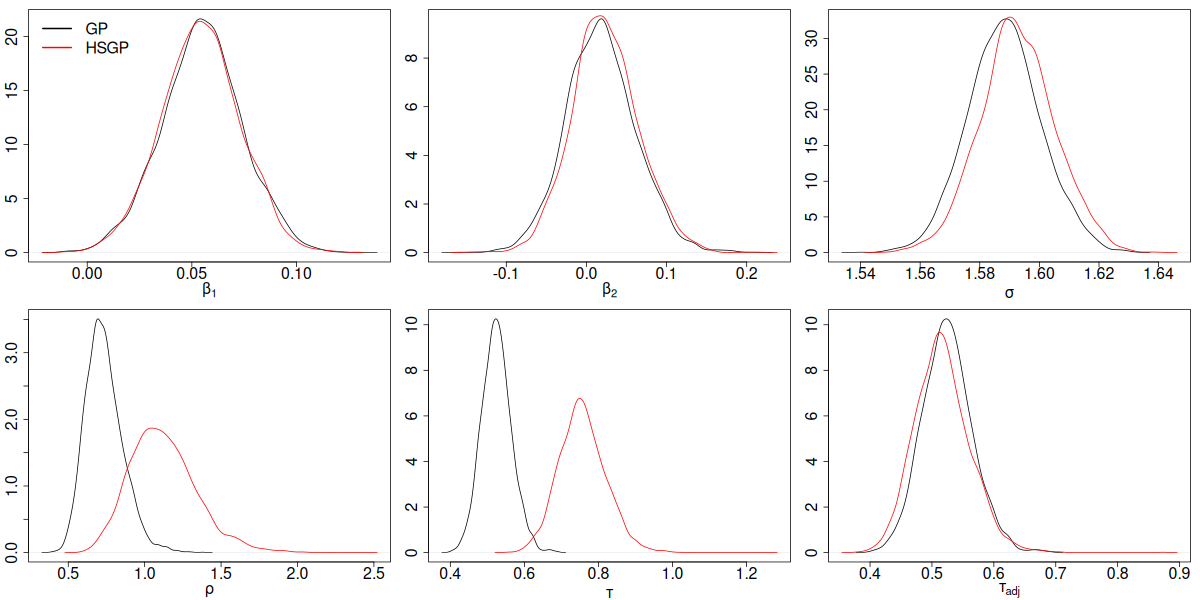
GP vs HSGP parameter posterior density
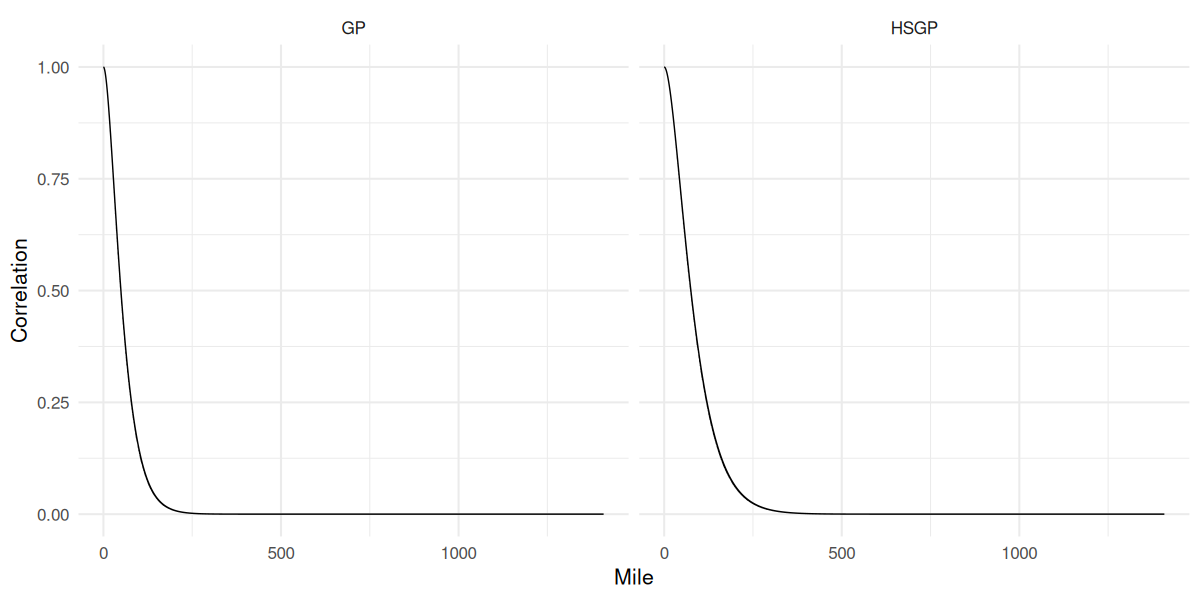
GP vs HSGP correlation function
GP vs HSGP effective sample size
Prepare for next class
- Work on HW 05 which is due Apr 8
- Complete reading to prepare for Tuesday’s lecture
- Tuesday’s lecture: Bayesian Clustering
References