loc_id hemoglobin anemia age urban LATNUM LONGNUM
1 1 12.5 not anemic 28 rural 0.220128 21.79508
2 1 12.6 not anemic 42 rural 0.220128 21.79508
3 1 13.3 not anemic 15 rural 0.220128 21.79508
4 1 12.9 not anemic 28 rural 0.220128 21.79508
5 1 10.4 mild 32 rural 0.220128 21.79508
6 1 12.2 not anemic 42 rural 0.220128 21.79508Scalable Gaussian Processes #1
Apr 01, 2025
Review of previous lectures
Two weeks ago, we learned about:
Gaussian processes, and
How to use Gaussian processes for
- longitudinal data
- geospatial data
Motivating dataset
Recall we worked with a dataset on women aged 15-49 sampled from the 2013-14 Democratic Republic of Congo (DRC) Demographic and Health Survey. Variables are:
loc_id: location id (i.e. survey cluster).hemoglobin: hemoglobin level (g/dL).anemia: anemia classifications.age: age in years.urban: urban vs. rural.LATNUM: latitude.LONGNUM: longitude.
Motivating dataset
Modeling goals:
Learn the associations between age and urbanicity and hemoglobin, accounting for unmeasured spatial confounders.
Create a predicted map of hemoglobin across the spatial surface controlling for age and urbanicity, with uncertainty quantification.
Map of the Sud-Kivu state
Last time, we focused on one state with ~500 observations at ~30 locations.
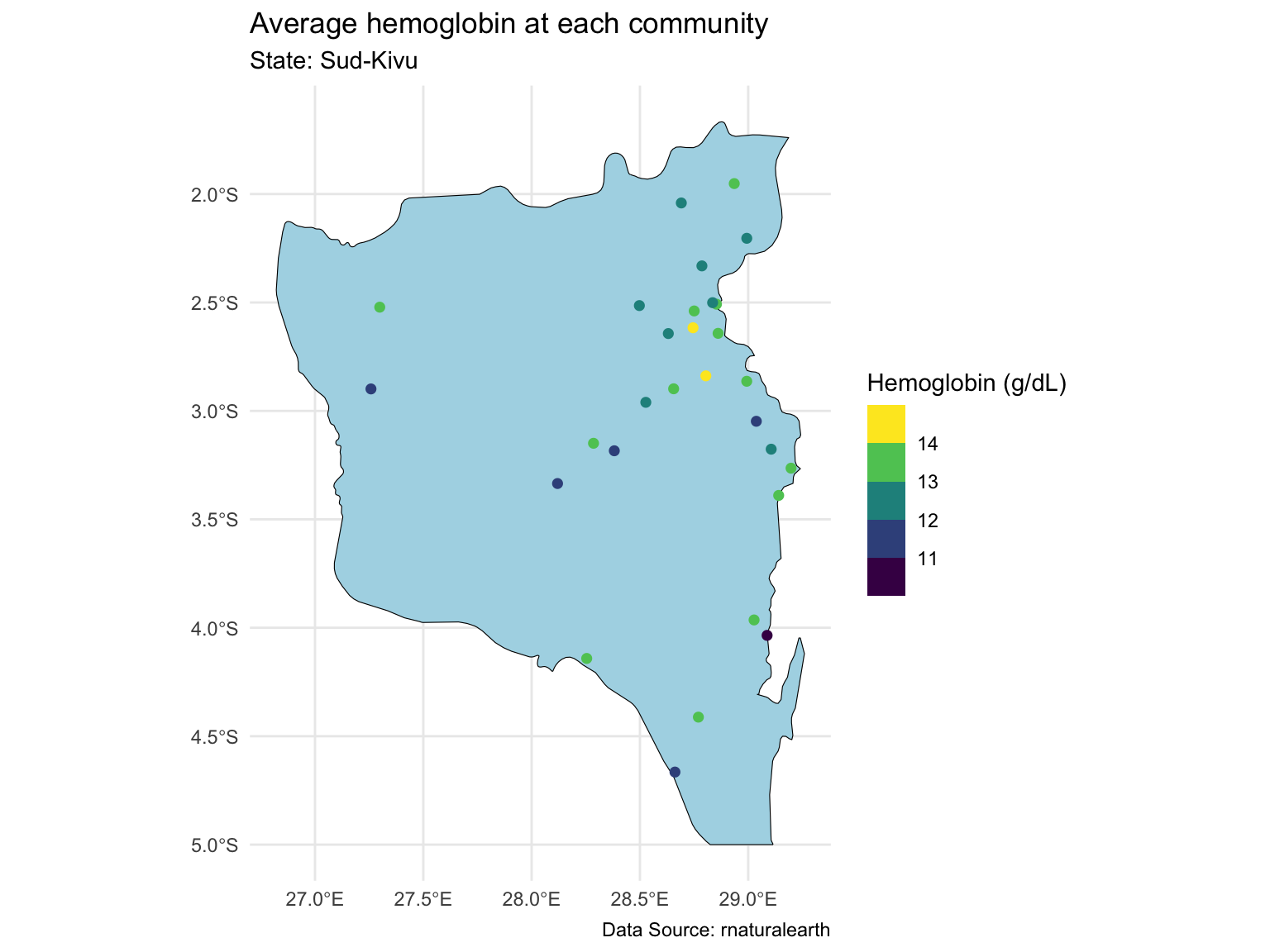
Prediction for the Sud-Kivu state
And we created a
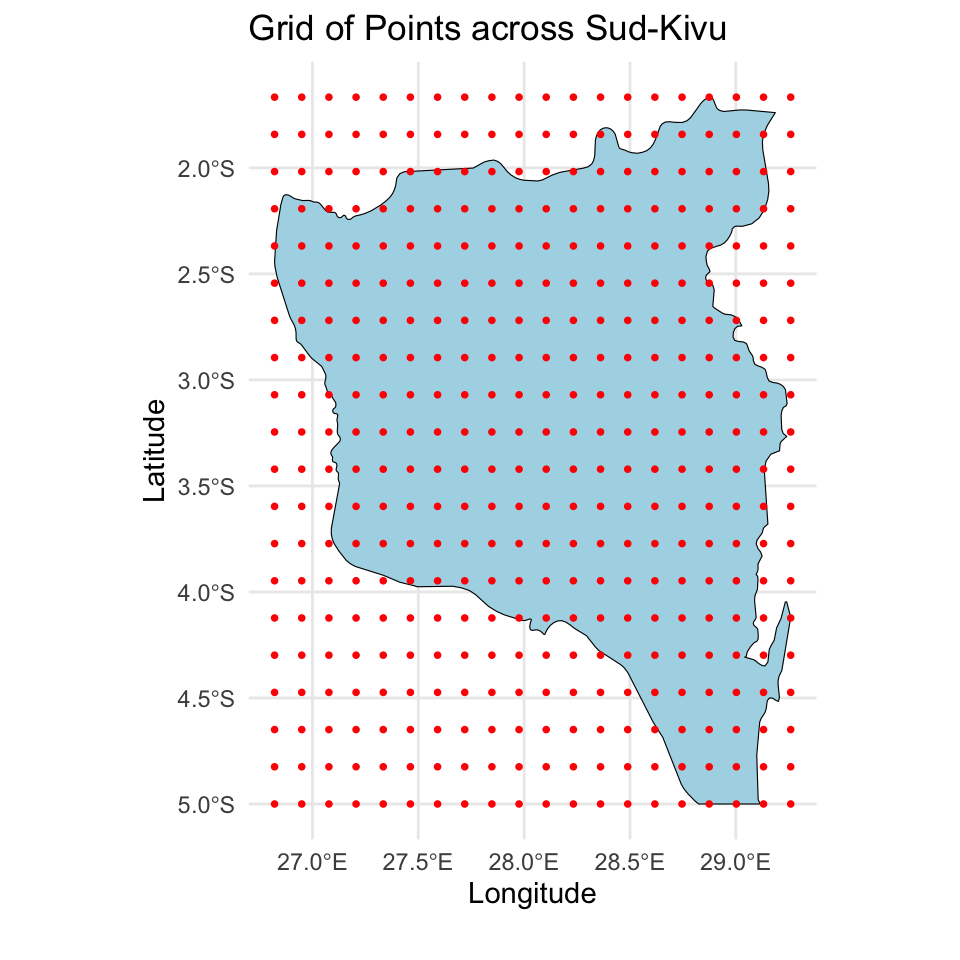
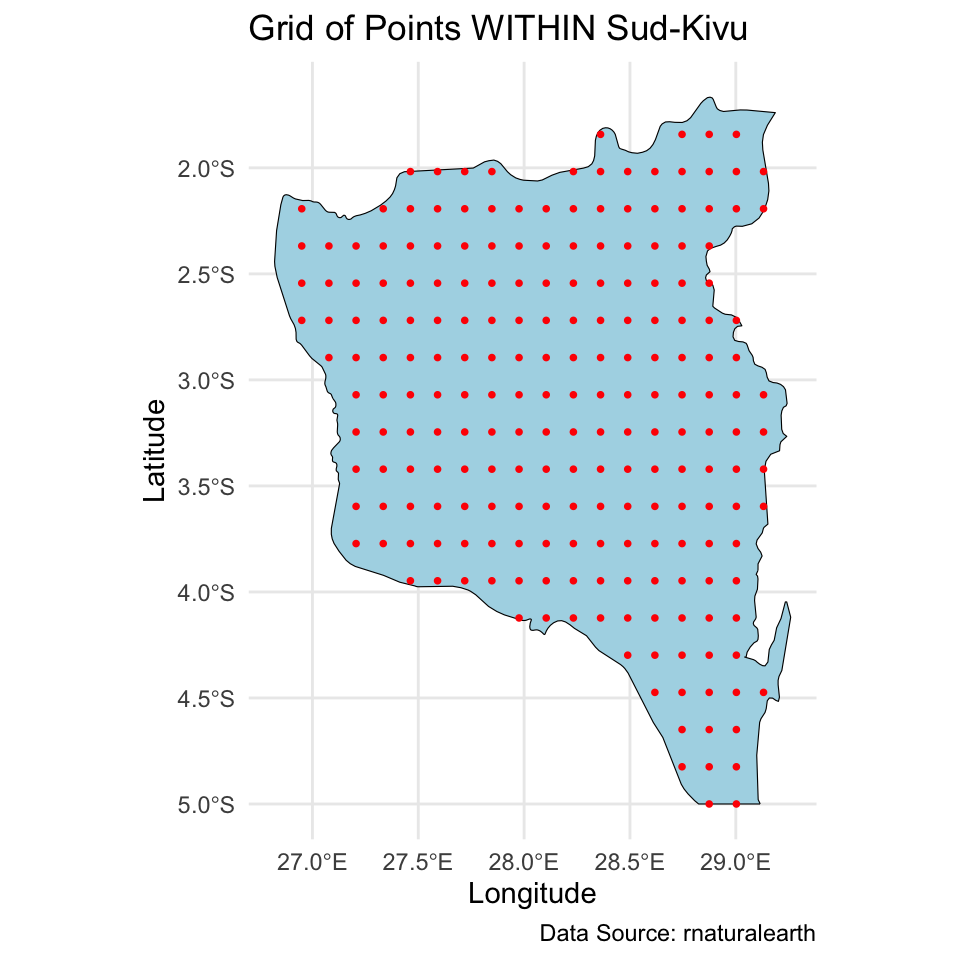
Map of the DRC
Today we will extend the analysis to the full dataset with ~8,600 observations at ~500 locations.
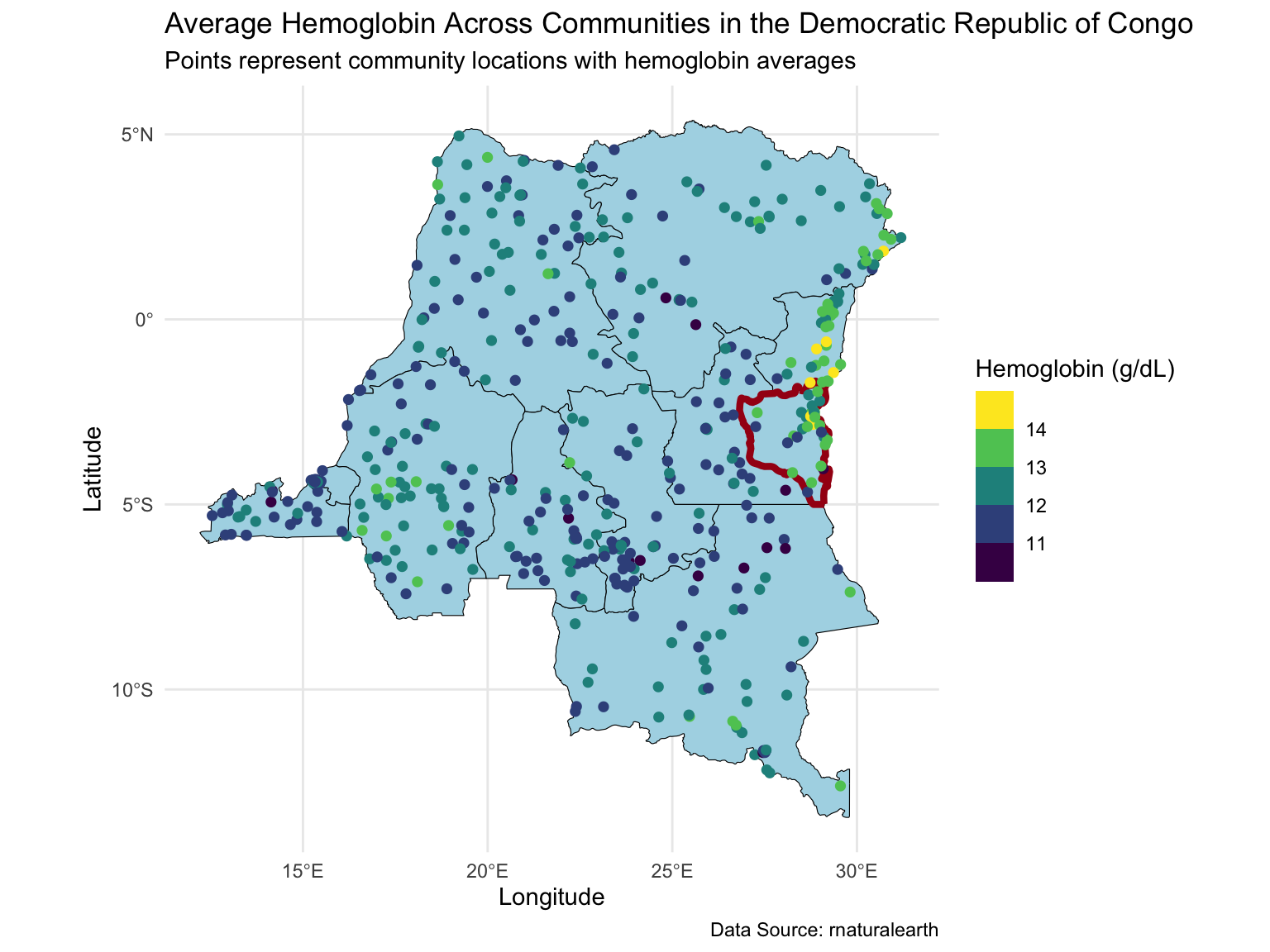
Prediction for the DRC
And we will make predictions on a
Modeling
Data objects:
Modeling
Population parameters:
Location-specific parameters:
Location-specific notation
Full data notation
Modeling
We specify the following model:
Computational issues with GP
Effectively, the prior for
This is not scalable because we need to invert an
Scalable GP methods overview
The computational issues motivated exploration in scalable GP methods. Existing scalable methods broadly fall under two categories.
Sparsity methods
Low-rank methods
Hilbert space method for GP
Lecture plan
Today:
- How does HSGP work
- Why HSGP is scalable
- How to use HSGP for Bayesian geospatial model fitting and posterior predictive sampling
Thursday:
- Parameter tuning for HSGP
- How to implement HSGP in
stan
HSGP approximation
Given:
- an isotropic covariance function
- a compact domain
HSGP approximates the
HSGP approximation
HSGP approximation
In matrix notation,
Why HSGP is scalable
- No matrix inversion.
- Each MCMC iteration requires
- Ideally
Model reparameterization
Under HSGP, approximately
Therefore we can reparameterize the model as
Note the resemblance to linear regression:
HSGP in stan
Similarly, we can use the reparameterized model in stan.
This is called the non-centered parameterization in stan documentation. It’s recommended for computational efficiency for hierarchical models.
transformed data {
matrix[n,m] PHI;
matrix[N,m] Z;
matrix[N,p] X_centered;
}
parameters {
real alpha_star;
real<lower=0> sigma;
vector[p] beta;
vector[m] b;
vector<lower=0>[m] sqrt_S;
...
}
model {
vector[n] theta = PHI * (sqrt_S .* b);
target += normal_lupdf(y | alpha_star + X_centered * beta + Z * theta, sigma);
target += normal_lupdf(b | 0, 1);
...
}Posterior predictive distribution
We want to make predictions for
Likelihood:
Kriging:
Posterior distribution:
Kriging
Recall under the GP prior,
where
Therefore by properties of multivariate normal,
Kriging under HSGP
Under HSGP,
Kriging under HSGP
Again by properties of multivariate normal,
- If
- But what if
Kriging under HSGP
If
By properties of multivariate normal,
Show if
Kriging under HSGP
Under the reparameterized model,
During MCMC sampling, we can obtain posterior predictive samples for
Kriging under HSGP – alternative view
Under the reparameterized model, there is another (perhaps more intuitive) way to recognize the kriging distribution under HSGP when
We model
HSGP kriging in stan
If stan.
If
Recap
HSGP is a low rank approximation method for GP.
- for covariance function
- on a box
- with
We have talked about:
- why HSGP is scalable.
- how to do posterior sampling and posterior predictive sampling in
stan.
HSGP parameters
Solin and Särkkä (2020) showed that HSGP approximation can be made arbitrarily accurate as
But how to choose:
- size of the box
- number of basis functions
Our goal:
Minimize the run time while maintaining reasonable approximation accuracy.
Note: we treat estimation of the GP magnitude parameter
Prepare for next class
Work on HW 05 which is due Apr 8
Complete reading to prepare for Thursday’s lecture
Thursday’s lecture:
- Parameter tuning for HSGP
- How to implement HSGP in
stan
References